Making of a new digital brain atlas FEM mesh
- 1. The itch
- 2. The challenges
- 3. The masterplan
- 4. The code
- 5. The revelation
1. The itch
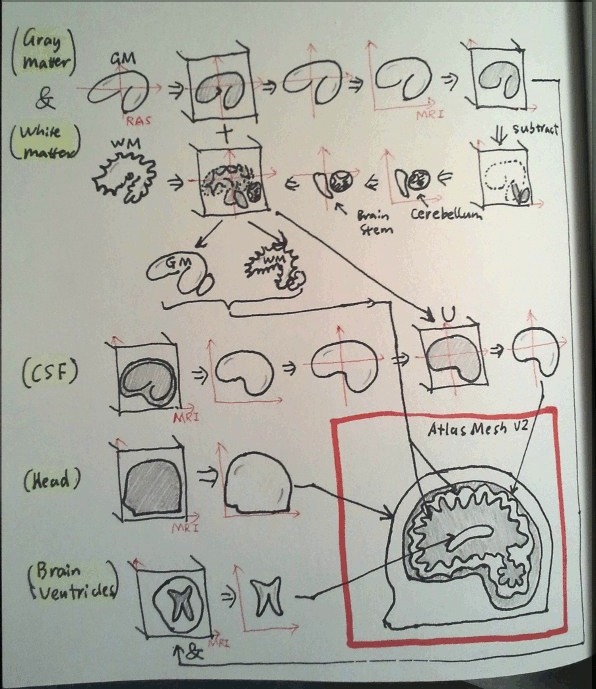
In the beginning, this brain atlas FEM mesh was created solely for the purposes of testing the Mesh-based Monte Carlo (MMC) code performance with a complex geometry, also making pretty figures for my paper. Because of the limited resolution of the segmented image and the constraints of the meshing toolbox at the time, the mesh was not anatomically accurate. However, soon after releasing the mesh to the public, I was approached by friends and colleagues, who have expressed their desires of using this mesh for their brain functional studies. In the same time, I heard the complains about the inaccuracy of the mesh, for example, the CSF layer is larger than expected, the gyri and sulci on the pial (gray-matter) surface is not so obvious, and so on. Surprisingly, an anatomically accurate brain atlas mesh is absent for the community, probably part of the reason is due to lack of meshing tools that are capable to handle complex volumetric images. After lots of searchings and readings, I was finally convinced that, after all, making an accurate brain atlas model is something worthwhile spending time with.
My desires of making such a mesh was further amplified by my continuous improvement of the iso2mesh toolbox, which is targeted at solving the exact problem, i.e. creating high-quality tetrahedral mesh from volumetric images. With a number of new meshing pipelines added to iso2mesh, the goal of making a better brain atlas mesh is getting more and more tangible.
2. The challenges
A number of issues must be solved in order to accomplish this task:
- obviously, to create an more accurate brain atlas mesh, particularly the gray and white matter surfaces, the MNI segmented brain volume of 1x1x1 mm resolution is not sufficient.
- FreeSurfer is a powerful tool to segment brain anatomy and create high-resolution gray-/white-matter surfaces. However, the FreeSurfer surfaces have multiple problems:
- they are extremely dense: each surface contains about 350000 nodes which is overkilling in some modeling tasks
- they typically contain self-intersecting triangular elements; these must be fixed before feeding to an mesh generator
- you can only get gray-/white-matter surfaces, no CSF and skin/skull
- they do not include cerebellum, brain stem and ventricles, unfortunately.
- the MNI segmented brain atlas do contain cerebellum, brain stem and ventricles, however, they are labeled with the same numbers as gray and white matters.
- the FreeSurfer surfaces are defined the RAS coordinate system, but the MNI image is in the MRI coordinate system.
3. The masterplan
4. The code
![[Home]](/img/mcx_wiki_banner.png)