Release Note for the Colin27 Brain Atlas FEM Mesh Version 2
the journey towards making an anatomically accurate digital brain FEM mesh
- by Qianqian Fang <fangq at nmr.mgh.harvard.edu>
- -- a quantitative analysis of the mesh generation method and results will be summarized in a full paper that is in preparation.
- 1. The inception
- 2. The challenges
- 3. The masterplan
- 4. The code
- 5. The afterthoughts
1. The inception
At the beginning, this brain atlas FEM mesh was created for nothing but testing the Mesh-based Monte Carlo (MMC) code performance with a complex geometry, as well as making pretty figures for my paper. Because of the limited image resolution and the constraints of the meshing tools at the time, the mesh was not anatomically accurate. Interestingly, soon after the first release of the mesh, I was approached by friends and colleagues, who showed their interests in using this mesh for their brain functional studies. In the meantime, the unsatisfactory about the inaccuracy of the mesh began to surface from these interactions, for example, the CSF layer is thicker than expected, the gyri and sulci on the pial (gray-matter) surface are not clear, and so on. Surprisingly, an anatomically accurate brain atlas mesh is absent for the community, probably part of the reason is due to lack of meshing tools that are capable of handling complex volumetric images. After extensive search and reading, I was finally convinced that, after all, making an accurate brain atlas model is something worthwhile doing.
My thought of making such a mesh was further amplified by my continuous improvement of the iso2mesh toolbox, which is targeted at solving the exact problem: creating high-quality tetrahedral mesh from volumetric images. With a number of new meshing pipelines added in iso2mesh, the goal of making a better brain atlas mesh was getting more and more tangible.
2. The challenges
To improve beyond the previous version, a number of issues must be considered and handled:
- Apparently, the MNI segmented brain volume is no longer sufficient to enhance the resolution and accuracy of the brain anatomy. This is particularly true for the gray matter (GM) and white matter (WM) surfaces. Additional inputs must be sought such as those from FreeSurfer.
- FreeSurfer is a powerful tool to segment brain anatomy and create high-resolution gray-/white-matter surfaces. However, the FreeSurfer surfaces posses multiple issues:
- they are extremely dense: each surface alone carries over 350000 nodes which is overkilling in many modeling tasks;
- they typically contain self-intersecting triangular elements; these must be fixed before feeding to a mesh generator;
- you can only get gray-/white-matter surfaces, no CSF and skin/skull;
- the surfaces only cover the cortex domain, they do not include cerebellum, brain stem and ventricles.
- The MNI segmented brain atlas do contain cerebellum, brain stem and ventricles, however, they are labeled with the same numbers as GM/WM. They must be separated and merged with FreeSurfer meshes to build a full-head mesh.
- The FreeSurfer surfaces are defined in the RAS coordinate system, but the MNI image is in the MRI coordinate system. These two must be registered before use simultaneously.
- We want to keep the total node number under a manageable size (preferably less than 100,000). That means the sum of all nodes on the surfaces of skin, CSF, GM and WM should be certainly below this limit.
For issue 2.1, the existing function "meshresample" is able to down-sample the surfaces nicely, however, it can not fix the self-intersecting elements (i.e. issue 2.2). Fortunately, the new additions "remeshsurf" and "surf2vol" in iso2mesh can be used to achieve both. For issue 2.3, we can exploit the gray-scale-image surface extraction feature in iso2mesh to produce a more accurate CSF and head surfaces. The more difficult part is related to issues 2.4/3. To "patch" the missing brain anatomies, one has to run multiple trips back and forth between the FreeSurfer surfaces and the MRI image space. To best preserve the features from the high-density GM/WM surfaces from FreeSurfer, we also need to use sub-millimeter resolution in the voxelation process. At the same time, we can not ignore issue 5, which requires us to keep the node number small. This is where the "radbound" option in iso2mesh comes in to play: one can use the radbound parameter to define the maximum element size for each given surface. Moreover, another parameter, "distbound", can be even more helpful here: it can enable iso2mesh (more precisely, CGAL) to produce fine mesh near sharp edges, and coarse mesh in smooth regions. A combination of the two should allow us to get close to the desired mesh size.
3. The masterplan
Before we layout a specific plan, we want to make some assumptions first to help us simplify the problem:
- Assumption 1: we assume that the 4 surfaces we are about to create - skin, CSF, pial and white-matter - are enclosing each other in the mentioned order. There is no intersection between the surfaces.
- Assumption 2: given the resolution of the images, we assume the minimum distance between any of the two surfaces is 2 mm (this can be smaller if we increase the mesh density)
Here comes the master-plan (inspired by the "master-plan" in the "Fantastic Mr. Fox"):
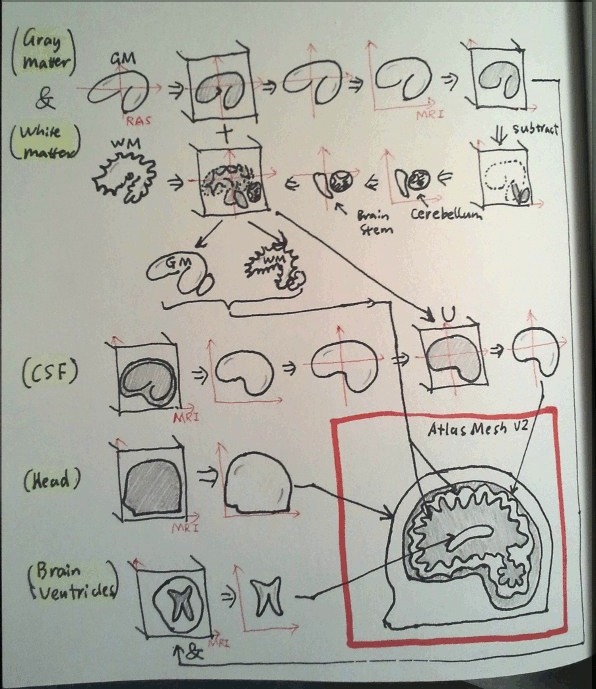
- First, we want to get the missing volumes for cerebellum and brain stem. To do so, we need to
- load FreeSurfer GM surface
- voxelate the surface to a volume in the RAS space
- extract the down-sampled surface
- map the down-sampled GM surface to the MRI space
- re-voxelate the GM surface in the MRI space
- mask and subtract the brain segmentation and get the volumes corresponding to the missing anatomy
- extract the surfaces for the GM/WM inside cerebellum/brain stem
- map the surfaces to the RAS space
- Second, we merge the GM/WM volumes and extract the surfaces. To do so, we
- load FreeSurfer WM surface
- voxelate the WM surface in the RAS space
- voxelate the cerebellum GM/WM surfaces in the RAS space, and merge with GM/WM volumes
- extract surfaces for GM/WM regions
- Third, we re-create the CSF surface in the high-resolution RAS space. We will
- convert the complete GM surface from RAS to MRI space
- voxelate the GM surface
- load the MRI image, union with the GM volume to get the CSF volume
- extract CSF surface in the MRI space
- Then, we re-create the skin surface. This is straightforward, simply extracting the surface from the MRI image
- The only thing left is the brain ventricles. We will
- convert the WM surface to the MRI space
- voxelate the surface, and mask out the ventricles from the MRI image
- extract surfaces from the ventricle images
- At last, we merge all 5 surfaces - skin, CSF, GM, WM, ventricles - into one surface, and call the mesh generator to populate tetrahedra inside each layer.
The following drawing shows graphically how the meshing process looks like
4. The code
The script, listed below, takes about half a hour on a relatively new machine, among which the first two "surf2vol" calls alone take about 15 min). This script also requires your system to have >6 GB memory. If you are interested in reproducing the results, please let me know and I can share you with the raw data files.
Anyway, here we go
%% improve Colin27 adult human brain atlas mesh using FreeSurfer surfaces, fangq, 2011/02/02-02/05 fprintf(1, '================================================ Defining the constants ====\n'); dim=[181 217 181]; offset=dim/2; dx=0.5; % the upper/lower limits consider the future added cerebellum and csf xi=-74:dx:74; yi=-92:dx:94; zi=-85:dx:74; mindist=2; SMOOTH=mindist/dx; THRESH=0.5; optpial.radbound=4; optpial.distbound=2; optwm.radbound=6; optwm.distbound=2; optcsf.radbound=8; optcsf.distbound=5; ISO2MESH_SESSION='atlasmesh_v2_';
%% loading the surfaces fprintf(1, '=========================================== Loading freesurfer surfaces ====\n'); [wln,wlf]=readasc('surf/lh.smoothwm.asc'); [wrn,wrf]=readasc('surf/rh.smoothwm.asc'); [pln,plf]=readasc('surf/lh.pial.asc'); [prn,prf]=readasc('surf/rh.pial.asc'); [pialn,pialf]=mergemesh(pln,plf,prn,prf); [wmn,wmf]=mergemesh(wln,wlf,wrn,wrf);
%% load the original MNI segmented brain fprintf(1, '============================================ Loading MNI segmented head ====\n'); fid=fopen('orig/atlas.img','rb'); head=fread(fid,inf,'uchar'); fclose(fid); head=reshape(head,dim); head(find(head>3))=4;
%% convert pial surface to a volume fprintf(1, '=========================================== Voxelating pial/wm surfaces ====\n'); %% one should either use the following 3 lines, or comment out them and use the 4th line pialv=surf2vol(pialn,pialf,xi,yi,zi); wmv=surf2vol(wmn,wmf,xi,yi,zi); save freesurfer_volumes pialv wmv % load freesurfer_volumes %% use load to avoid repetitive meshing of pial and wm wmvfill=fillholes3d(wmv,0); pialvfill=fillholes3d(pialv,0); clear pialv wmv
%% let pial surface enclosing the white matter surface fprintf(1, '=============================================== Extracting pial surface ====\n'); pialvfill=pialvfill | thickenbinvol(wmvfill,2);
%% convert binary image to gray scale to create smooth surface pialvfillsmooth=smoothbinvol(pialvfill,SMOOTH);
%% extract an intersecting-element-free pial surface from the volume [pialns,pialfs]=v2s(pialvfillsmooth,0.48,optpial);
%% conver the new surface back to the original coordinate system fprintf(1, '================================ Maping pial surface to the image space ====\n'); pialns1=pialns*dx; pialns1=pialns1+repmat([xi(1),yi(1),zi(1)]+dx,size(pialns1,1),1);
%% now we need to add cerebellum gray and white matters
%% conver the pial surfaces from RAS to volume coordinates and convert to volume mask fprintf(1, '=============================================== Voxelating pial surface ====\n'); pialns1(:,1)=-pialns1(:,1); pialns1=pialns1+repmat(offset,size(pialns1,1),1); fsgmvol=surf2vol(pialns1,pialfs(:,1:3),-1:offset(1)*2-2,-1:offset(2)*2-2,-1:1:offset(3)*2-2); fsvol=fillholes3d(fsgmvol,0);
%% diffvol stores the original image - freesurfer volume = cerebellum volume fprintf(1, '========================================= Get cerebellum and brain stem ====\n'); idx=find(head==4 | head==1); diffvol=head; diffvol(idx)=0; idx=find(fsvol>0); diffvol(idx)=0;
%% mesh the cerebellum gray and white matters fprintf(1, '============================== Meshing gray/white matters in cerebellum ====\n'); cbwm=(diffvol==3); cbgm=(diffvol>0) | thickenbinvol(cbwm,2); cbgmsmooth=smoothbinvol(cbgm,SMOOTH); cbwmsmooth=smoothbinvol(cbwm,SMOOTH); clear cbgm cbwm [cbgmn,cbgmf]=v2s(cbgmsmooth,THRESH,3); [cbwmn,cbwmf]=v2s(cbwmsmooth,THRESH,3); cbwmn2=sms(cbwmn,cbwmf(:,1:3),SMOOTH,THRESH,'lowpass'); cbgmn2=sms(cbgmn,cbgmf(:,1:3),SMOOTH,THRESH,'lowpass'); clear cbgmsmooth cbwmsmooth cbwmn cbgmn %[cbn,cbf]=mergemesh(cbwmn2,cbwmf(:,1:3),cbgmn2,cbgmf(:,1:3)) %[nodet,elemt,facet]=s2m(cbn,cbf,1,100);
%% convert the cerebellum surfaces to the RAS system fprintf(1, '=============================== Converting cerebellum mesh to RAS space ====\n'); cbgmn2ras=cbgmn2-repmat(offset,size(cbgmn2,1),1); cbgmn2ras(:,1)=-cbgmn2ras(:,1); cbwmn2ras=cbwmn2-repmat(offset,size(cbwmn2,1),1); cbwmn2ras(:,1)=-cbwmn2ras(:,1);
%% convert the cerebellum surfaces to volume in the RAS space cbgmnv=surf2vol(cbgmn2ras,cbgmf,xi,yi,zi); cbwmnv=surf2vol(cbwmn2ras,cbwmf,xi,yi,zi); cbwmnvfill=fillholes3d(cbwmnv,0); cbgmnvfill=fillholes3d(cbgmnv,0); clear cbwmnv cbgmnv cbgmn2ras cbwmn2ras
%% merge cerebellum gm/wm with the fore-brain gm/wm volumes, respectively fprintf(1, '============================== Merging cerebellum with voxelated FS mesh ====\n'); wmvfill=wmvfill | cbwmnvfill; pialvfill=pialvfill | cbgmnvfill | thickenbinvol(wmvfill,SMOOTH); clear cbwmnvfill cbgmnvfill
%% extract surfaces from the merged white and gray matters pialvfillsmooth=smoothbinvol(pialvfill,SMOOTH); wmvfillsmooth=smoothbinvol(wmvfill,SMOOTH); fprintf(1, '================================= Extracting merged pial and wm surfaces ====\n'); [pialns,pialfs]=v2s(pialvfillsmooth,THRESH-0.1,optpial); [wmns,wmfs]=v2s(wmvfillsmooth,THRESH+0.1,optwm); clear pialvfillsmooth wmvfillsmooth
%% convert to the correct coordinates pialns=pialns*dx; pialns=pialns+repmat([xi(1),yi(1),zi(1)]+dx,size(pialns,1),1); wmns=wmns*dx; wmns=wmns+repmat([xi(1),yi(1),zi(1)]+dx,size(wmns,1),1);
%% extract CSF layer from the original image fprintf(1, '================================= Extracting CSF volume from image space ====\n'); csfv=head; csfv(head==4)=0; csfv=(csfv>0); csfv=fillholes3d(csfv,0); csfvsmooth=smoothbinvol(csfv,SMOOTH);
%% get CSF surface [csfn,csff]=v2s(csfvsmooth,THRESH,4); csfn2=sms(csfn,csff(:,1:3),4,THRESH,'lowpass');
%% convert CSF surface from image space to RAS space fprintf(1, '======================================= Mapping CSF surface to RAS space ====\n'); csfn2ras=csfn2-repmat(offset,size(csfn2,1),1); csfn2ras(:,1)=-csfn2ras(:,1); fprintf(1, '================================================= Voxelating CSF surface ====\n');
%% convert CSF surface to a volume in the RAS space csfnv=surf2vol(csfn2ras,csff(:,1:3),xi,yi,zi); csfnvfill=fillholes3d(csfnv,0); csfrasv=csfnvfill | thickenbinvol(pialvfill,SMOOTH); csfrasvsmooth=smoothbinvol(csfrasv,SMOOTH);
%% extract the CSF surface from the volume fprintf(1, '================================================= Extracting CSF surface ====\n'); [csfrasn,csfrasf]=v2s(csfrasvsmooth,THRESH-0.1,optcsf); csfrasn=csfrasn*dx; csfrasn=csfrasn+repmat([xi(1),yi(1),zi(1)]+dx,size(csfrasn,1),1);
%% smooth the generated surfaces fprintf(1, '================================================== Smoothing all surfaces ====\n'); pialns1=sms(pialns,pialfs(:,1:3),SMOOTH,THRESH,'lowpass'); wmns1=sms(wmns,wmfs(:,1:3),SMOOTH,THRESH,'lowpass'); csfrasn1=sms(csfrasn,csfrasf(:,1:3),SMOOTH,THRESH,'lowpass'); fprintf(1, '================================= Extracting skin surface from MNI image ====\n');
%% load Colins atlas V1 mesh, extract the head surface, and % merge with new surfaces headsmooth=smoothbinvol(head>0,SMOOTH); [headn,headf]=v2s(headsmooth,THRESH,8); headn1=sms(headn,headf(:,1:3),SMOOTH,THRESH,'lowpass');
%% convert surfaces to the volumetric coordinates fprintf(1, '============================= Converting all RAS surfaces to image space ====\n'); pialns1(:,1)=-pialns1(:,1); pialns1=pialns1+repmat(offset,size(pialns1,1),1); wmns1(:,1)=-wmns1(:,1); wmns1=wmns1+repmat(offset,size(wmns1,1),1); csfrasn1(:,1)=-csfrasn1(:,1); csfrasn1=csfrasn1+repmat(offset,size(csfrasn1,1),1);
%% extract the ventricle surface from MNI atlas inside the freesurfer white matter fprintf(1, '================================= Masking ventricles and extracting mesh ====\n'); fswmvol=surf2vol(wmns1,wmfs(:,1:3),-1:offset(1)*2-2,-1:offset(2)*2-2,-1:1:offset(3)*2-2); venv=(head==1) & fillholes3d(fswmvol,0); venv=thinbinvol(venv,1); venvsmooth=smoothbinvol(venv,2); [venn,venf]=v2s(venvsmooth,0.5,2); venn1=sms(venn,venf(:,1:3),4,0.5,'lowpass');
%% combine all surfaces, transform to the volume coordinates fprintf(1, '=================================================== Merging all surfaces ====\n'); [colinsn,colinsf]=mergemesh(headn1,headf(:,1:3),csfrasn1,csfrasf(:,1:3),... pialns1,pialfs(:,1:3),wmns1,wmfs(:,1:3),venn1,venf(:,1:3));
%% define mesh density metric propotional to z-coordinates fprintf(1, '=================================================== Defining density map ====\n'); hdsize=(1-(headn1(:,3)-min(headn1(:,3)))/(max(headn1(:,3))-min(headn1(:,3))))*8+2; cssize=(1-(csfrasn1(:,3)-min(csfrasn1(:,3)))/(max(csfrasn1(:,3))-min(csfrasn1(:,3))))*5+2; gmsize=(1-(pialns1(:,3)-min(pialns1(:,3)))/(max(pialns1(:,3))-min(pialns1(:,3))))*4+2; wmsize=(1-(wmns1(:,3)-min(wmns1(:,3)))/(max(wmns1(:,3))-min(wmns1(:,3))))*5+2; vnsize=(1-(venn1(:,3)-min(venn1(:,3)))/(max(venn1(:,3))-min(venn1(:,3))))*4+2;
%% add the mesh density metric as the last column of the node array colinsn(:,end+1)=[hdsize;cssize;gmsize;wmsize;vnsize];
%% define internal points for each anatomic region p1=[90 50 132]; p2=[90 50 127]; p3=[94 94 130]; p4=[111 99 135]; p5=[131 87 90]; regions=[p1;p2;p3;p4;p5]; fprintf(1, '==================================================== Making the 3D mesh ====\n');
%% do the meshing [node,elem,face]=surf2mesh(colinsn,colinsf,min(colinsn(:,1:3))-1,max(colinsn(:,1:3))+1,1,100,regions,[]); save final_mesh
%% final packaging fprintf(1, '========================================= Packaging the data to release ====\n'); fc=finddisconnsurf(face(:,1:3)); headsurf=fc{1}; csfsurf=[fc{2};fc{4};fc{6}]; gmsurf=fc{3}; wmsurf=fc{5}; face=[headsurf ones(size(headsurf,1),1); csfsurf 2*ones(size(csfsurf,1),1); ... gmsurf 3*ones(size(gmsurf,1),1); wmsurf 4*ones(size(wmsurf,1),1)]; elem(find(elem(:,end)==6),5)=5; elem(find(elem(:,end)==1),5)=3; elem(:,end)=elem(:,end)-1; elem(:,1:4)=meshreorient(node(:,1:3),elem(:,1:4)); face(:,1:3)=meshreorient(node(:,1:3),face(:,1:3)); README_brain_mesh=sprintf(['Collins adult brain atlas FEM mesh - Version 2L (low-resolution).\n\n' ... 'Created on 02/05/2011 by Qianqian Fang [1] with iso2mesh [2] version 1.0.0.\n' ... 'The gray/white matter surfaces were created by Katherine Perdue [3] with FreeSurfer [4]\n\n' ... 'Please refer to ''Qianqian Fang, "Mesh-based Monte Carlo method using fast ray-tracing in Plucker ' ... 'coordinates," Biomed. Opt. Express 1(1), 165-175 (2010)'' for details.\n\n' ... 'Format: \n' ... ' node: node coordinates (in mm)\n' ... ' face: surface triangles, the last column is the surface ID, \n' ... ' 1-scalp, 2-CSF, 3-gray matter, 4-white matter\n' ... ' elem: tetrahedral elements, the last column is the region ID, \n' ... ' 1-scalp and skull layer, 2-CSF, 3-gray matter, 4-white matter\n\n' ... 'URL: http://mcx.sourceforge.net/cgi-bin/index.cgi?MMC/CollinsAtlasMesh\n\n' ... 'References:\n [1] http://nmr.mgh.harvard.edu/~fangq/\n [2] http://iso2mesh.sf.net\n'... ' [3] Email: kperdue@nmr.mgh.harvard.edu\n [4] http://surfer.nmr.mgh.harvard.edu']); save MMC_Collins_Atlas_Mesh_Version_2L.mat node elem face README_brain_mesh fprintf(1, '============================================ The masterplan has succeeded ====\n');
5. The afterthoughts
Some of you may get scared when reading the complex diagram and script in the preceding sections. I am sure you are not alone. After all, it is not a simple problem we are dealing with. We are trying to build something quite complex, not complex per se, but depending on imperfect and non-coordinating results from several other components, not mention we have a list of constraints to satisfy. On a second though, I feel quite excited. In the end, the script "magically" did the work, automatically! The magic rather comes from the ability to access each incremental step over a complex meshing pipeline, facilitated by the modular design of the iso2mesh toolbox. This is exactly what this toolbox is meant to be: it is not only a one-liner mesh generator, but also it exposes you all the internal modules so you can combine them and use them intelligently to solve much more complex problems.
I am going to put my week-long excitement to a rest, and I want to share the experience with you by writing this "emotional" journal. Hopefully, your fun of creating meshes has just started ...
![[Home]](/img/mcx_wiki_banner.png)